Author(s): Viraj Mehta
Glioblastoma multiforme is a deadly brain cancer with a median patient survival time of 18-24 months. A single biopsy cannot provide complete assessment of the tumor’s microenvironment, making personalized care limited. 50% of the patients do not respond to the anti-cancer drug Temozolomide(TMZ) because of the over-expression of MGMT gene. Epigenetic silencing of the MGMT gene by methylation results in decreased MGMT expression, resulting in increased sensitivity to TMZ, and longer survival. The purpose of this research is to use artificial intelligence (AI) to design a low cost methodology to determine the MGMT’s methylation status and suggest non- invasive treatment plan.
Introduction
Glioblastoma multiforme is a very lethal form of brain cancer
with no known cure. The prognosis remains poor with a 5%
average survival rate, despite aggressive treatments. For high grade
gliomas, treatment combines surgical resection, postoperative
radiation, and chemotherapy using temozolomide (TMZ), an
alkylating agent [1]. Although radiation therapy and chemotherapy
with TMZ contribute to lengthen the survival and improve quality
of life, the survival advantages are still palliative due to TMZ
resistance as primary reason for GBM treatment failure as 50%
of TMZ treated patients don?t respond to TMZ [2].
The methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) gene promoter region impacts sensitivity to temozolomide by repairing the main cytotoxic lethal base pairs, which are composed of the alkylating agent TMZ, and hence, is linked to reliably predicting effectiveness of TMZ.
MGMT is an enzyme involved in DNA dealkylation and mediation of DNA damage, and is overexpressed in 60% of glioblastomas [1]. MGMT encodes for a DNA repair enzyme that provides resistance to alkylating chemotherapies such as temozolomide (TMZ). Because MGMT transcription can be silenced by promoter methylation in tumor cells, MGMT promoter methylation in patient tumors causes decreased MGMT protein expression, thereby abrogating the DNA repair activity necessary for TMZ resistance (mechanism explained in the Figure 1(a) below) [3]. Thus, patients with a methylated MGMT promoter have improved survival and better response to radiation with concurrent temozolomide-based therapy. Hence, methylation of the O-6-methylguanine-DNA methyltransferase (MGMT) gene promoter has Emerged as a strong prognostic factor for newly diagnosed glioblastoma [3].
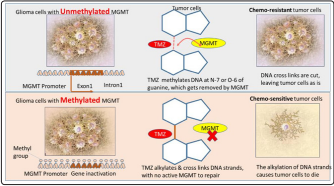
Figure 1(a): Chemosensitivity of MGMT: Enhanced due to
epigenetic silencing of the DNA-repair gene MGMT [10].
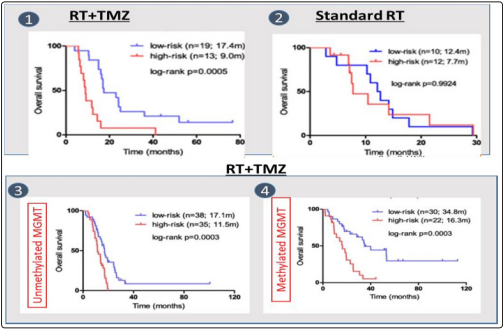
The picture below shows survival rate for low risk and high risk patients in 4 cases: (1): The survival improvement for patients with radio therapy (RT) using temozolomide (2); standard radiotherapy without temozolomide(3) impact of RT+TMZ in case of unmethylated MGMT
(4) Imapct of RT+TMZ in case of methylated MGMT (4) on the survival rate for low-risk and high-risk patients is shown below in Figure 2. Radiotherapy and TMZ combined confers a clear overall survival benefit to patients with methylated MGMT relative to standard radiotherapy. Ref: Novel predictive epigenetic signature for TMZ in glioblastomas Additionally, MGMT can be epigenetically repressed in multiple ways, most commonly due to methylation of its promoter region, or by over-expression of several microRNAs [4].
MicroRNAs are small noncoding RNAs that target specific sequences of mRNAs, thereby regulating gene expression, causing translational repression or mRNA degradation [4].
This is summarized in the following table:
Problem Statement
Current methods for tumor detection and MGMT gene methylation
identification in order to recommend effective TMZ dosages to
glioblastoma patients are extremely invasive and time- and cost-
intensive. Current methods use invasive biopsies and expensive
procedures like genetic testing that add to the financial burden
and unnecessary time lag between patient testing and therapy
recommendation.
Proposal
Train a convolutional neural networks and the combined TCGA
and TCIA data to predict MGMT methylation status and overall
survival for patients with methylated and unmethylated MGMT.
Perform computation modeling using miRNA databases and
MGMT interaction to find miRNAs that can regulate MGMT
expression, leading to improved TMZ sensitivity for patients with
unmethylated MGMT.
Methods and Materials
Tools and framework used: Python 3, OpenCV, TensorFlow
Tumor segmentation using the Convolutional Neural Network Architecture
Neural network on the TCIA dataset provides evaluation of state-of-the-art methods for the segmentation of intrinsically heterogeneous (in appearance, shape, and histology) brain tumors, namely gliomas. A CNN was trained on the BraTS dataset to create a pixel-wise segmentation map that would predict the tumor region in a brain MRI scan. The Convolutional Neural Network contains an encoder path, which reduces image dimensionality to capture the context of the image, and a decoder path, which enables precise localization using transposed convolutions and up-sampling.
It uses a binary mask to classify each pixel in the image as either containing or not containing a tumor region. The CNN architecture uses convolutional layers and pooling layers. The number of channels is shown on top of each box. The white boxes represent feature maps and arrows indicate different operations (concatenation, convolution, pooling etc).
The Sorenson-Dice coeffcient was used to predict accuracy: the Sorensen?Dice coefficient is a statistic used for comparing the similarity of two samples, defined as
Dice (P, T) = 2|p ? T|, where P is algorithmic predictions ? {0,1} and T is consensus truth ? {0, 1} |P|+|T|
CNN model applied to TCIA patient dataset for tumor segmentation
CNN (Convolutional Neural Network) model trained on BraTS dataset above resulted in high accuracy. The trained CNN was then applied (run) on the TCIA dataset to detect tumor areas.
Genomics (methylation data) was retrieved from the TCGA website and filtered by TCGA- GBM, DNA methylation, and methylation ? value [5]. Methylation values for these 3 mentioned sites were taken, and a patient was considered to have a positive MGMT methylation status if the maximum of the three was greater than a specific threshold. In general, the methylation value for each site is expressed as a ? value, representing a continuous measurement from 0 (completely unmethylated) to 1 (completely methylated). Following the merge of the preprocessed T1 modality MRIs from the TCIA dataset with the methylation label, the resulting methylation data was mapped to patient IDs and T1 images. The data was then used to train a second CNN to predict a patient?s methylation status.
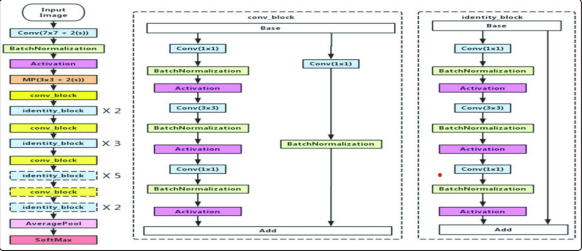
In general, in a deep convolutional neural network, several layers are stacked and trained with labeled data. The network then learns features at various levels of abstraction (high/medium/low level). In residual learning, instead of trying to learn features directly, it tries to learn from a residual. CNN does this by using shortcut connections (directly connecting input of nth layer to some (n + x)th layer.
The CNN classification model has 5 stages each with a Convolution and Identity block. Each convolution and identity block has 3 convolutional layers. Each image frame was first input in a CNN layer, and multiple residual convolutional layers were used to detect edges/shapes/corners that represent the tumor in the image.
For patients with ?unmethylated MGMT,? computational modeling was done to find miRNAs and binding strength with MGMT. Binding sites for miRNAs in the 3? UTR of MGMT were predicted. In addition, stronger miRNA-mediated repression of mRNA was observed with multiple binding sites for the same miRNA on 3?UTR.
| Gene = MGMT miRNA | Geometric Share Complementarity Score | Duplex structure | MFE kcal/mol (min free energy) |
|---|---|---|---|
| miR181d-5p | 142 | miRNA 3? ugggUGGCUGUUGUUACUUACAa 5? || |:| | |:|||:||| Target 5? taaaACAGGCCA-AGTGAGTGTg 3? | -20.9 |
| miR767 | 139 | miRNA 3? ucuuuGGUACCCCAUACUCGUcu 5? :||| ||||||||| Target 5? tggctTCAT---GTATGAGCAag 3? | -13.50 |
| miR603 | 158 | miRNA 3? ugCGCGU-CCGGUC---UCU----- GGGUCCGu 5? |: || |||||| ||: ||||||| Target 5? ctGTCCAGGGCCAGCTAAGGCCCATCCC AGGCc 3? | -24.80 |
| miR-409-3p | 125 | miRNA 3? uccCCAAGUGGCUC-- GUUGUAag 5? | | ::||||| |:|||| Target 5? caaGCTCTGCCGAGGCCGACATga 3? | -13.3 |
| miR124-3p | 135 | miRNA 3? ccguaagUGGCG--CACGGAAu 5? :|||| ||||:|| Target 5? tgtgcgaGCCGCGAGTGCTTTc 3? | -15.6 |
Results
1) Overall survival: Kaplan Meier curve, drawn using Xenabrowser, demonstrates higher survival for patients with methylated MGMT
vs unmethylated MGMT (drawn for a subset of the total dataset of patients). Ref: xenabrowser.net
2) miRNA-MGMT mRNA analysis: In reference to Table 2, high absolute values of Minimum Free Energy and GSC (Geometric
Share Complementarity Score) demonstrate the stability of the miR181d and miR603 binding. Thus, miR603 and miR181d can
silence MGMT and improve glioma cell sensitivity to TMZ.
The regression analysis between miRNA and MGMT mRNA from the TCGA dataset indicated that miR-603 and miR-181d are consistently inversely related with MGMT mRNA in the TCGA dataset (p value of 0.005 and 0.006 respectively).
| miR-181d-5p | miR-603 | miR-409-3p | miR-124-3p | |
|---|---|---|---|---|
| r | -0.218 | -0.211 | 0.073 | -0.011 |
| p-value | 0.006 | 0.005 | 0.363 | 0.882 |
Discussion
The first CNN model used for tumor segmentation was a fully
convolutional model with an encoder-decoder architecture. The
CNN consists of 4 down-sampling blocks, in which the input
image is scaled down using convolutional layers, followed by 4
up-sampling blocks in which the input image is scaled up using
transpose convolution blocks.

The second CNN, a classification model uses two fully connected layers at the end to classify each image into one of two categories, methylated or unmethylated. The CNN has residual connections that enable an easier propagation of the error into the lower layers of the network. Additionally, the loss used was a categorical cross entropy loss with acategorical cross entropy accuracy for evaluation.
Categorical cross-entropy loss is defined as

Where i=1 if methylation is false, i=2 if methylation is true, f (si):
probability output for class i
ti: 1 for true ground truth class, 0 otherwise. Simply put, it is the
(negative) log of the probability output for the ground truth class.
Conclusion
Gene methylation is a control mechanism that regulates gene
expression. MGMT is a DNA repair enzyme that causes resistance
to the effect of alkylating chemotherapy. Aberrant methylation
deactivates the gene, leading to loss of MGMT protein expression
and reduced proficiency to repair DNA damage induced by
alkylating chemotherapeutic agents, thereby increasing tumor
susceptibility to alkylating agent-based chemotherapy [6-8].
References
1. Daniel Chow, Peter Chang, Brent D. Weinberg, Daniela
A. Bota, Jack Grinband, et al. (2008) ?Imaging Genetic
Heterogeneity in Glioblastoma and Other Glial Tumors:
Review of Current Methods and Future Directions: American
Journal of Roentgenology (AJR).? American Journal of
Roentgenology 210: 30-38.
2. Sang Y Lee (2016) ?Temozolomide Resistance in Glioblastoma
Multiforme.? Genes & Diseases, Elsevier 3: 198-210.
3. Brandes Alba A (2017) ?Role of MGMT Methylation
Status at Time of Diagnosis and Recurrence for Patients
with Glioblastoma: Clinical Implications.? The Oncologist,
AlphaMed Press 22: 432-437.
4. Cabrini Giulio (2015) ?Regulation of Expression of O6-
Methylguanine-DNA Methyltransferase and the Treatment of
Glioblastoma (Review).? International Journal of Oncology,
D.A. Spandidos 47: 417-428.
5. GDC. (n.d.). Retrieved November 08, 2020, from https://
portal.gdc.cancer.gov/
6. Chang P (2018) ?Deep-Learning Convolutional Neural
Networks Accurately Classify Genetic Mutations in Gliomas.?
American Journal of Neuroradiology, American Journal of
Neuroradiology 39: 1-7.
7. Hegi Monika E (2005) ?MGMT Gene Silencing and Benefit
from Temozolomide in Glioblastoma: NEJM.? New England
Journal of Medicine 352: 997-1003.
8. Coverage Policies and Guidelines.? Medical Policy Manual |
BlueCross BlueShield of Tennessee, http://www.bcbst.com/
providers/coverage-policies-guidelines/index.page.
