Author(s): Ajayi AO*, Owaboriaye M, Rufus J, Arowolo HM and Osunla CA
Background: Pathogenic viruses and bacteria of bat origin causes a lot of diseases that often leads to epidemics and pandemics. Fruit bats were documented reservoir of many viral infections including COVID-19. This study was designed to detect the presence of some enteric viruses of gastroenteritis as well as bacterial species from Eidolon helvum bat species in Nigeria.
Methodology: Twenty-five samples were collected between January and March 2018, from six different locations in Ondo State, Nigeria. The samples were tested for the presence of Rotavirus and Adenovirus antigens using the CerTest Biotech 4th generation Quadruple Enzyme Immunoassay (EIA) kit according to the manufacturers’ specification. The bacterial species were isolated by pour plating and identified using standard microbiological methodsand API kits.
Results: Throughout the period of this study, two samples (10%) were tested positive for both Norovirus and Adenovirus in bat fecal samples from Epinmin Akoko, Nigeria, while the isolated bacterial species from bats in Owo included Streptococcus Spp, Micrococcus Spp, Staphylococcus aureus, Enterococcus Spp., Staphylococcus epidermidis, Salmonella typhi, Echerichia coli, Klebsiella oxytoca, Acinetobacter iwoffi, Citrobacter freundii, Serratia marcescens, Enterobacter cloacae, Proteus vulgaris, Enterobacter agglomerans and Proteus mirabillis.
Conclusion: The isolation of these microbes are suggestive of infectious agents that can be of threat to public health and caution bush meat consumers from erratic consumption of unverified bush meat culinaries. The data obtained from this study is valuable for public health management as well as disease prevention and control.
Bats (order Chiroptera) are a diverse group of mammals found on every continent of the world except the Polar Regions and a few oceanic islands. There are 1150 species of bats in the world, meaning that one in five (21 percent) mammal species is a bat. Bats are distinguished from other mammals by their evolution of true flight, as opposed to the gliding capabilities of mammals in other orders. Bats are mammals whose forelimbs have been adapted to form webbed wings, making them the only mammals naturally capable of true and sustained flight. The taxonomy of bats is currently being revised in the light of molecular phylogenetics, as they have traditionally been split into two suborders - Microchiroptera and Megachiroptera, based largely on the evolution of echo-location (ability to navigate using the reflection of sound waves). Bats are the second largest order of mammals (after the rodents), with about 1,240 bat species with the less specialized and largely fruit-eating megabats, or flying foxes, and the highly specialized and echolocating microbats which are potential reservoirs of many infectious diseases [1-9]. Noroviruses (NoVs) of the family Caliciviridae are one of the viruses being transmitted by bats. Noroviruses are small nonenveloped viruses approximately 27-35 nm in diameter with a positive-sense, single stranded RNA genome that were first discovered in human outbreak of gastroenteritis in Norwalk, Ohio, which gave the name Norwalk virus (NV) to the prototype strain of NoV, in 1968. NoV infection occurs in the small intestine although the replication may not be restricted to the enterocytes. Epidemiological studies have repeatedly shown spread of this virus in human populations, while serological prevalence of 22.1% was found in laboratory mice in North America. Norovirus infects a wide range of animals including dairy cattle in The Netherlands, veal calves in the US, In Germany and Japan as well as 4-weekold lion cub in Pistoia zoo in Italy [10-20]. The virus is infectious at very low doses and transmits rapidly by the feco-oral route via contaminated food and water, causing a severe, sporadic or more than 85% of epidemic diarrhea and vomiting in all age groups, especially during the winter. In a previous hospital based study in Lagos, the prevalence of norovirus diarrhea was 37.3%, while reported a prevalence of 25.5% in a community based study in Osun state, Nigeria [22, 32, 33].
Members of the Adenoviridae family are non-enveloped, icosahedral viruses, of about 70-90nm in size, with a linear, doublestranded DNA genome ranging from 26 to 46 kb. DNA sequence analysis indicates that this family is divided into five genera: Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus and Ichtadenovirus. Adenoviruses (AdVs) infect a wide range of vertebrates, including mammals, birds, amphibians, reptiles and fish. Members of the Mastadenovirus and Aviadenovirus genera only infect mammals and birds [24,25]. Recently, great attention has been paid to bats given their role as reservoirs of emerging viruses, including SARS coronavirus, Ebolavirus, Hendra virus and Nipah virus [6]. Acute infective gastroenteritis is a major global health problem, which manifests as three or more watery or loose bowel evacuations with fever and vomiting in a 24-hour period that may last several days. Children under 5 years of age are particularly susceptible, and global estimates indicated a mean of between 3.5 and 7 episodes of severe diarrhea during the first 2 years of life, and the greatest burden is in the developing countries because of poor sanitation, lack of safe drinking water, and bad sanitary habits. Several studies in Nigeria showed that more than 315,000 deaths of preschool age children are recorded annually as a result of diarrhea disease humans include Group-A rotaviruses, astroviruses, human caliciviruses and the enteric adenoviruses. Rotaviruses are members of the family Reoviridae and genus Rotavirus and contain 3 primary species: Rotavirus A, Rotavirus B, and Rotavirus C. The diagnostic system is also applicable to other viral groups [26-35].
The most widely used method for laboratory diagnosis is the enzyme immunoassay (EIA or ELISA), which detects the RVA group antigen (VP6) directly on stools. The genetic methods - microarray and real-time PCR (RT-qPCR) - are very sensitive and capable of discriminating mixed RVA infections, and have been successfully developed and employed in diagnosis of RV (34). The bacterial population of bat feacal dropping was also described by Wolkers et al., whose study emphasized that bats are commonly regarded as vectors for viruses, but paucity of information is available about bacterial communities in bats and the possible role of bats in the transmission of foodborne diseases. The reported bacterial genera predominantly recovered from fecal bacteria in bats were found to be diverse with Corynebacterium, Serratia, Pseudomonas, Enterococcus and Yersinia. The opportunistic bacterial pathogens including Citrobacter freundii, Escherichia coli, Enterococcus faecalis, Serratia fonticola and Rahnella aquatilis were also obtained [8, 36]. The microbial species detected in bat feacal droppings in this study can cause acute diarrheal disease (ADD) amongst other infectious diseases that can result to diseases outbreaks. Acute diarrheal disease (ADD) is a syndrome caused by different agents (bacteria, viruses and parasites), and its major manifestation is increased number of bowel movements, with watery or loose stools in the susceptible host [5,8,12]. This study therefore helps to elucidate the involvement of different microbial groups that can be harbored by bats. This will help to control infection from these sources.
The study site used in Ondo State, Nigeria were based on the availability of Bat colonies in this area as listed here. This includes, Akure, Epinmi-Akoko, Ikare-Akoko, Supare-Akoko, Ugbe-Akoko as well as on a roof top of an uncompleted building at Owo, Ondo state Nigeria for intensive enteric virus screening and bacteriological analysis analysis.
Between January to March 2013, during each visit to the bat colonies, early in the morning. Bat fecal samples were collected from five different locations (Ikare, Supare, Ugbe, Epinmi and Akure) in Ondo State. Freshly voided samples were aseptically collected into sample bottles and preserved with ice packs/blocks in a clean cooler before transportation to the laboratory for analysis.
The fecal samples were tested for the presence of Rotavirus and Adenovirus antigens using the Certest Biotech Quadriple EIA kit according to the manufacturer’s specification (Certest, 2013).
The strip consists of a nitrocellulose membrane pre coated with mouse monoclonal antibodies on the test line (T), in the result window, against Rotavirus or Adenovirus and with rabbit polyclonal antibodies on the control line (C) against a specific protein. The label/sample absorbent pad is sprayed with labelled solution (mouse monoclonal antibodies anti-Rotavirus) conjugated to red polystyrene latex and control label solution (Specific binding protein) conjugated to green polystyrene latex, forming coloured conjugate complexes.
Sample was mixed properly with the Extraction buffer to ensure good sample dispersion and extraction of the virus antigen. The Certest kit was removed from its sealed bag and four drops of the homogenized sample was introduced into the window marked with letter A and B. The results were taken at ten minutes. Materials used for this study includes sample bottles, 70% of Ethanol, cotton wool, nose cover, hand gloves, cardboard, CerTest Biotech Quadruple EIA kit for the detection of adenovirus and rotavirus, cooler for the preservation of samples and kits used. Similarly, the bacterial species were identified using standard microbiological methods including API kits.
Eosine methylene blue agar, peptone water and Nutrient agar were generally used for the bacteriological analysis. 1gram of the sample was weighed and was diluted in 9ml sterile peptone water and serial dilution was done .1ml of inoculum was taken from 10- 1 ,10-2,10-3 dilution to seed sterile plates of Eosine methylene blue (EMB) agar and Nutrient agar in replicates employing pour plate technique An un-inoculated plate serve as the growth control for the two culture plates. (Nutrient agar and EMB agar). The plates were swirled for even saturation of the inoculum and the media, the plates were properly labelled, inverted and incubated at 37°C for 24hrs. After 24hrs, the result colonies were carefully examined and determined for its cultural characteristics, the diversity and proportion of the isolates. The Gram negative organisms were identified using API kit.
The API-20E fast identification system combines some of conventional tests and allows the identification of a limited number of Gram negative Enterobacteriaceae or Non-Enterobacteriaceae. The test system is stored in 20 small reaction tubes or wells containing dehydrated substrate to detect enzymatic activity, usually related to fermentation of carbonhydrates or catabolism of proteins or amino acids by the inoculated organisms. A bacterial suspension is used to rehydrate each of the wells and the strips are incubated. During incubation, metabolism produces colour changes that are either spontaneous or revealed by the additionof reagents. All positive and negative test results are compiled to obtain a profile numbers in a commercial codebook or manual to determine the identification of the bacterial isolates.
1. ONPG: Test for β-galactosidase enzyme by hydrolysis of the
substrate o-nitrophenyl-b-D-galactopyranoside
2. ADH: Decarboxylation of the amino acid arginine by arginine
dihydrolase
3. LDC: Decarboxylations of the amino acid lysine by lysine
decarboxylase
4. ODC: Decarboxylations of the amino acid ornithine by
ornithine decarboxylase
5. CIT: Utilization of citrate as only carbon source
6. H2S: Production of hydrogen sulfide
7. URE: Test for the enzyme urease
8. TDA (Tryptophan deaminase): Detection of the enzyme
tryptophan deaminase: Reagent to put- Ferric Chloride.
9. IND: Indole Test-production of indole from tryptophan by
the enzyme tryptophanase. Reagent10. VP: The Voges-Proskauer test for the detection of acetoin
(acetyl methylcarbinol) produced by fermentation of glucose
by bacteria utilizing the butylene glycol pathway
11. GEL: Test for the production of the enzyme gelatinase which
liquefies gelatin
12. GLU: Fermentation of glucose (hexose sugar)
13. MAN: Fermentation of mannose (hexose sugar)
14. INO: Fermentation of inositol (cyclic polyalcohol)
15. SOR: Fermentation of sorbitol (alcohol sugar)
16. RHA: Fermentation of rhamnose (methyl pentose sugar)
17. SAC: Fermentation of sucrose (disaccharide)
18. MEL: Fermentation of melibiose (disaccharide)
19. AMY: Fermentation of amygdalin (glycoside)
20. ARA: Fermentation of arabinose (pentose sugar)
Suspension of each isolates were prepared in a sterile distilled water and it was introduced into the API20E biochemical test strip which contains dehydrated bacterial bio-chemical reagents in 20 separate compartments accordingly with a sterile syringe. Sterile oil was added into the Arginine dihydrolase (ADH), Lysine decarboxylase (LDC), Ornithine decarboxylase (ODC), Hydrogen sulphide (H2S), and Urease (URE) compartments, Some drops of water is added in the tray and the API test strip was placed on the tray and it was incubated for 24 hours.
The colour change was observed after 24 hours, reagents were added to some specific compartments. One drop of ferric chloride was added to Tryptophan deaminase (TDA), one drop of Kovacs reagent was added to indole (IND) and one drop of KOH (VP reagent1 and reagent 2) was added to Voges-Proskauer (VP) and the result was gotten after 10minutes. The organism was identified by using API catalog.
The test was performed to determine the phenotypic resistance of the bacterial isolates to commonly used antibiotics. This test was carried out following the Kirby-Bauer disk diffusion methods of CLSI, (37). Inoculum from culture of bacteria isolates on nutrient agar slants were inoculated into test tubes containing sterilized nutrient broth and incubated at 37 for 18hours which serve as the stock for the test.1ml from this stock was then transferred into sterilized petri plate and 20ml of already prepared and sterilized Mueller Hinton agar was inoculated. The plates were then allowed to cool for about 15 minutes so as to allow the inoculated plate to gel and excess surface moisture to be absorbed before applying the antibiotics impregnated discs. Predetermined commercial Gram negative and gram positive discs were applied to the surface of the well labelled inoculated agar plates aseptically using sterile forceps. The disks were placed firmly by slightly pressing on the inoculated plates with the sterilized forceps to ensure complete contact with the agar, the discs were applied according. After 18hours of incubation, each plate was examined. The susceptibility responses of the isolates to respective antibiotics were indicated by a clear zone of inhibition measured using a calibrated ruler which was held on the back of the inverted petri-plate and was recorded. The isolates were then scored as either sensitive to the antibiotics or resistant depending on the diameter of the zone of inhibition.
Figure 1 shows the map of the study area in Ondo State, Nigeria, from where this bat dropping samples were collected. The specific areas with bat colonies includes Owo in Owo Local Government for the bacterial species analysis, Akure, Akure South Local Government Area; Epinmi in Akoko South East Local Government Area, Ikare in Akoko North East Local Government Area, Supare in Akoko South West Local Government Area and Ugbe in Akoko South East Local Government Area, Ondo State manily for the viral species analysis.
Throughout the period of this study, two samples tested positive for both Adenovirus and Norovirus in bat fecal samples from Epinmin in Akoko South East Local Government Area, Ondo State, Nigeria (Tables 1 and 2). Between January and March, 2018, a total number of twenty samples were collected from the five locations and tested. The fecal characteristics and result of the test are presented in Tables 1, 2 and Fig. 2. Throughout the period of this study, two samples tested positive for both Norovirus and Adenovirus in bat fecal samples from Epinmin in Akoko South East Local Government Area, Ondo State, Nigeria.
| Specimencollection date | Location | Sample code | Time ofcollection | Time of analysis | Stoolcharacteristics | Norovirus |
|---|---|---|---|---|---|---|
| 20/01/2018 | Ikare | A | 7:00am | 8:00am | Solid and dry | - |
| 21/01/2018 | Ikare | B | 8:00am | 9:00am | Solid and dry | - |
| 23/01/2018 | Supare | C | 8:00am | 9:00am | Solid and dry | - |
| 24/01/2018 | Ugbe | D | 9:00am | 10:00am | Solid and dry | - |
| 26/01/2018 | Ugbe | E | 9:00am | 10:00am | Semi-solid | - |
| 31/01/2018 | Epinmi | F | 10:00am | 11:00am | Semi-solid | - |
| 01/02/2018 | Epinmi | G | 10:00am | 11:00am | Semi-solid | + |
| 02/02/2018 | Epinmi | H | 10:00am | 11:00am | Semi-solid | - |
| 03/02/2018 | Epinmi | I | 10:00am | 11:00am | Semi-solid | - |
| 04/02/2018 | Epinmi | J | 09:00am | 10:00am | Semi-solid | - |
| 05/02/2018 | Epinmi | K | 09:00am | 10:00am | Semi-solid | - |
| 06/02/2018 | Epinmi | L | 09:00am | 10:00am | Semi-solid | - |
| 07/02/2018 | Akure | M | 10:00am | 11:00am | Semi-solid | - |
| 08/02/2018 | Akure | N | 10:00am | 11:00am | Semi-solid | - |
| 09/02/2018 | Akure | O | 10:00am | 11:00am | Semi-solid | - |
| 10/02/2018 | Akure | P | 10:00am | 11:00am | Semi-solid | - |
| 11/02/2018 | Akure | Q | 10:00am | 11:00am | Semi-solid | - |
| 12/02/2018 | Akure | R | 10:00am | 11:00am | Semi-solid | - |
| 15/02/2018 | Akure | S | 10:00am | 11:00am | Semi-solid | - |
| Specimencollection date | Location | Sampleindicator | Time ofcollection | Time ofanalysis | Stoolcharacteristics | Adenovirus | Rotavirus |
|---|---|---|---|---|---|---|---|
| 0/01/2018 | Ikare | A | 7:00am | 8:00am | Solid and dry | - | - |
| 21/01/2018 | Ikare | B | 8:00am | 9:00am | Solid and dry | - | - |
| 23/01/2018 | Supare | C | 8:00am | 9:00am | Solid and dry | - | - |
| 24/01/2018 | Ugbe | D | 9:00am | 10:00am | Solid and dry | - | - |
| 26/01/2018 | Ugbe | E | 9:00am | 10:00am | Semi-solid | - | - |
| 31/01/2018 | Epinmi | F | 10:00am | 11:00am | Semi-solid | - | - |
| 01/02/2018 | Epinmi | G | 10:00am | 11:00am | Semi-solid | + | - |
| 02/02/2018 | Epinmi | H | 10:00am | 11:00am | Semi-solid | - | - |
| 03/02/2018 | Epinmi | I | 10:00am | 11:00am | Semi-solid | - | - |
| 04/02/2018 | Epinmi | J | 09:00am | 10:00am | Semi-solid | - | - |
| 05/02/2018 | Epinmi | K | 09:00am | 10:00am | Semi-solid | + | - |
| 06/02/2018 | Epinmi | L | 09:00am | 10:00am | Semi-solid | - | - |
| 07/02/2018 | Akure | M | 10:00am | 11:00am | Semi-solid | - | - |
| 08/02/2018 | Akure | N | 10:00am | 11:00am | Semi-solid | - | - |
| 09/02/2018 | Akure | O | 10:00am | 11:00am | Semi-solid | - | - |
| 10/02/2018 | Akure | P | 10:00am | 11:00am | Semi-solid | - | - |
| 11/02/2018 | Akure | Q | 10:00am | 11:00am | Semi-solid | - | - |
| 12/02/2018 | Akure | R | 10:00am | 11:00am | Semi-solid | - | - |
| 15/02/2018 | Akure | S | 10:00am | 11:00am | Semi-solid | - | - |
| 16/02/2018 | Akure | T | 10:00am | 11:00am | Semi-solid | - | - |
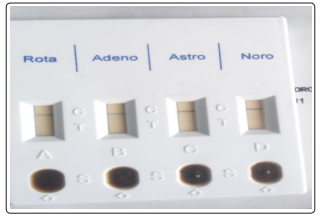
Figure 2: Sample of Certest Kit Used For Adenovirus, Norovirus and Rotavirus Examination in Faecal Sample Collected From Sample sources
The Gram positive bacteria recovered from fecal pellets of bat belongs to five genera namely: Streptococcus spp, Micrococcus spp, Staphylococcus Aureus, Enterococcus spp. and Staphylococcus Epidermidis [5]. Similarly, the Gram negative bacteria recovered from fecal pellets of bat belongs to ten genera namely: Salmonella Typhi, Echerichia Coli, Klebsiella Oxytoca, Acinetobacter Iwoffi, Citrobacter Freundii, Serratia Marcescens, Enterobacter Cloacae, Proteus Vulgaris, Enterobacter Agglomerans and Proteus Mirabillis (Tables 3 to 5) [10].
| Cultural characteristics.Isolates edge pigment shape | Cellular morphology | |||||
|---|---|---|---|---|---|---|
| Elevation | Morphology | Gram | ||||
| M1 | Pink | irregular | Rough | Flat | Cocci | G +ve |
| M2 | Milky | Circular | Rough | Flat | Rods | G - ve |
| M3 | Milky | Circuler | Rough | Flat | Tiny Cocci | G + ve |
| M4 | Milky | Irregular | Crenate | Raised | Rods | G - ve |
| M5 | Pink | Circular | Crenate | Flat | Rods | G - ve |
| M6 | Creamy | Irregular | Crenate | Flat | Cocci + Rods | G - ve |
| M7 | Pink | Irregular | Rough | Flat | Cocci | G + ve |
| M8 | Milky | irregular | Rough | Flat | Cocci | G + ve |
| M9 | Milky | Circuler | Crenate | Flat | Cocci | G + ve |
| M10 | Pink | Irregular | Crenate | Flat | Rods | G - ve |
| M11 | Milky | Irregular | Rough | Flat | Rods | G - ve |
| M12 | Creamy | Irregular | Rough | Raised | Rods | G - ve |
| M13 | Milky | Circular | Smooth | Raised | Cocci | G + ve |
| M14 | Pink | Irregular | Lobate | Raised | Rods | G - ve |
| M15 | Pink | Irregular | Smooth | Raised | Rods | G - ve |
| M16 | Pink | Circular | Smooth | Raised | Rods | G - ve |
| M17 | Milky | Circular | Rough | Flat | Tiny Cocci | G + ve |
| M18 | Creamy | Circular | Smooth | Raised | Rods | G - ve |
| M19 | Pink | Circular | Lobate | Raised | Rods | G - ve |
| M20 | Milky | Irregular | Rough | Flat | Rods | G - ve |
| Isolate | Catalase | Coagulase | Spore test | Oxidase | Haemolysin test | Glucose | Sucrose | Lactose | Probable organism |
|---|---|---|---|---|---|---|---|---|---|
| M1 | - | - | - | - | + | + NG | - NG | + NG | Streptococcus spp. |
| M3 | + | - | + | - | - NG | + GP | + GP | Micrococcus spp | |
| M7 | - | - | - | - | + | + NG | - NG | + NG | Streptococcus spp. |
| M8 | + | + | - | + | + | + NG | - GP | + GP | Staphylococcus aureus |
| M9 | - | + | - | - | - | + | - | - | Enterococcus spp. |
| M13 | + | - | - | - | - | + | - | + | Staphylococcus epidermidis |
| M17 | + | - | - | + | - | - NG | + GP | + GP | Micrococcus spp. |
KEY: NG= No Gas Production, GP= Gas Production
| Isolates | API identification code | Probable Organism | % probability |
|---|---|---|---|
| M2 | 4404540 | Salmonella typhi | 99.96 |
| M4 | 7542001 | Echerichia coli | 98.97 |
| M5 | 5445673 | Klebsiella oxytoca | 99.5 |
| M6 | 6306042 | Acinetobacter iwoffi | 95.3 |
| M10 | 5215772 | Klebsiella pneumonia | 99.87 |
| M11 | 3634552 | Citrobacter freundii | 94.1 |
| M12 | 7532000 | Echerichia coli | 80.58 |
| M14 | 7312153 | Serratia marcescens | 95.64 |
| M15 | 3305572 | Enterobacter cloacae | 71.47 |
| M16 | 2756000 | Proteus vulgaris | 81.06 |
| M18 | 5315572 | Enterobacter agglomerans | 97.76 |
| M19 | 2756400 | Proteus mirabillis | 99.43 |
| M20 | 7512000 | E. coli (inactive) | 76.82 |
The bacterial isolates showed multiple antibiotic resistant to antibiotics provided, with Levofloxacin and Ampiclox having high susceptibility effect against Gram positive bacteria. Similarly, Ofloxacin and Nalidixic acid having high susceptibility effect against Gram negative bacteria (Table 6A and B).
| Zones of inhibition by the antibiotics(MM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| ISOLATES | CIP | CIN | AMX | STR | LVX | ERY | CHL | APX |
| M1 | 12R | 10R | 0R | 5R | 10R | 10R | 15S | 16S |
| M2 | 13R | 14R | 0R | 4R | 15S | 8R | 10R | 17S |
| M3 | 13R | 12R | 6R | 3R | 13R | 8R | 18S | 18S |
| M4 | 10R | 10R | 9R | 0R | 13R | 0R | 12R | 20S |
| M5 | 13R | 15S | 0R | 0R | 15S | 12R | 13R | 13R |
| M6 | 13R | 13R | 6R | 0R | 12R | 13R | 14R | 14R |
| M7 | 11R | 15S | 4R | 6R | 10R | 14R | 15S | 15S |
| M8 | 14R | 10R | 0R | 8R | 14R | 15S | 16S | 18S |
| M9 | 15S | 12R | 5R | 10R | 13R | 15S | 15S | 19S |
| M10 | 10R | 11R | 6R | 12R | 15S | 8R | 8R | 18S |
| M11 | 14R | 8R | 0R | 9R | 18S | 10R | 6R | 15S |
| M12 | 13R | 9R | 8R | 10R | 16S | 13R | 13R | 13R |
| M13 | 10R | 12R | 0R | 8R | 18S | 6R | 6R | 14R |
| M14 | 12R | 13R | 6R | 9R | 15S | 15S | 15S | 14R |
| M15 | 14R | 15S | 8R | 11R | 10R | 18S | 8R | 15S |
| M16 | 10R | 13R | 9R | 5R | 0R | 6R | 16S | 20S |
| M17 | 11R | 12R | 0R | 8R | 0R | 12R | 17S | 15S |
| M18 | 13R | 11R | 5R | 0R | 16S | 8R | 8R | 20S |
| M19 | 12R | 10R | 8R | 0R | 15S | 10R | 15S | 20S |
| M20 | 14R | 8R | 0R | 11R | 10R | 10R | 15S | 0R |
CIP= Ciprofloxacin, CIN= cinoxacin, AMX= Amoxicillin STR= Streptomycin, LVX= levofloxacin, ERY= Erythromycin, CHL=
Chloramphenicol, APX= Ampiclox,
R-Resistant
S- Sensitive
| ISOLATES | Zones of inhibition by the antibiotics (MM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OFX | PEF | CPX | AUG | CIN | GEN | CEP | NAL | SXT | PEN | |
| M1 | 23S | 10R | 0R | 0R | 13R | 13R | 10R | 21S | 0R | 0R |
| M2 | 26S | 0R | 19S | 0R | 10R | 18S | 12R | 22S | 19S | 0R |
| M3 | 30S | 9R | 0R | 0R | 0R | 13R | 10R | 0R | 15S | 0R |
| M4 | 36S | 16S | 0R | 0R | 9R | 13R | 0R | 34S | 16S | 7R |
| M5 | 36S | 10R | 0R | 0R | 16S | 14R | 10R | 18S | 14R | 8R |
| M6 | 17S | 11R | 0R | 0R | 10R | 13R | 10R | 22S | 20S | 6R |
| M7 | 30S | 16S | 0R | 0R | 11R | 15S | 18S | 22S | 20S | 5R |
| M8 | 33S | 38R | 0R | 0R | 16S | 14R | 0R | 35S | 21S | 0R |
| M9 | 44S | 0R | 24S | 0R | 38S | 13R | 0R | 0R | 18S | 0R |
| M10 | 28R | 19S | 0R | 0R | 0R | 16S | 6R | 22S | 18S | 0R |
| M11 | 42S | 0R | 0R | 0R | 17S | 14R | 10R | 0R | 10R | 0R |
| M12 | 43S | 9R | 0R | 0R | 18S | 14R | 11R | 0R | 8R | 0R |
| M13 | 0R | 0R | 0R | 0R | 0R | 15S | 16S | 24S | 15S | 0R |
| M14 | 28S | 16S | 0R | 0R | 9R | 15S | 0R | 18R | 18S | 0R |
| M15 | 42S | 10R | 0R | 0R | 0R | 0R | 0R | 15S | 15S | 0R |
| M16 | 43S | 11R | 0R | 0R | 16S | 15S | 0R | 20S | 15S | 0R |
| M17 | 0R | 10R | 0R | 0R | 15S | 16S | 0R | 21S | 14R | 0R |
| M18 | 34S | 16S | 0R | 0R | 0R | 13R | 15S | 22S | 14R | 0R |
| M19 | 0R | 0R | 0R | 0R | 10R | 14R | 18S | 35S | 14R | 0R |
| M20 | 26S | 14R | 0R | 0R | 19S | 15S | 10R | 0R | 6R | 0R |
OFX= Ofloxacin, PEF= Pefloxacin, CPX= Ciprofloxacin, AUG= Augmentin, CIN= cinoxacin, GEN= Gentamycin, CEP=
Cephalosporine, NAL= Nalidixic acid,
SXT= Trimethopri-sulphamethoxazole PEN= Penicillin
R-Resistant
S-Sensitive
The global diversity of viruses found in bats is still largely unknown, and it is thought that a great number of virus taxa are yet to be discovered. Bats are often asymptomatic during infection, but the disease can be severe for a new host species when cross-species transmission occurs. Surveys of the bat virome and monitoring of virus dynamics in bat populations can inform our fundamental understanding on how viral pathogens emerge and evolve, enabling strategies for the prevention of future outbreaks or pandemics [38-40]. This present study shows the presence of Adenovirus in fecal bat from Epinmi, Ondo state. Metagenomic studies in bats from other countries have identified adenovirus, papillomavirus, parvovirus and a calicivirus (sapovirus), reported by [27, 41-47]. It is difficult to make specific or direct comparisons with the present study due to significant methodological variation among the studies including: sample type (i.e. fresh guano, urine, roost guano). Perhaps the most comparable study is that of, who used a metagenomics approach to examine virus diversity in African straw coloured fruit bats (Eidolon helvum) [41]. Human adenoviruses (HAdV) are major causes of clinical infections including respiratory diseases, gastroenteritis, and conjunctivitis [16]. Human Adenovirus belongs to the genus Mastadenovirus of the family Adenoviridae, The virion consists of non-enveloped, linear, dsDNA genome (26-45 kb) encapsidated in an icosahedral
protein shell. There are 52 serotypes classified into six species, A-G, of which species F serotypes 40 and 41 primarily affect the gut, contributing 5%-20% of hospitalizations for childhood diarrhea. The incidence of infection is nearly 3 times greater in developing countries than developed ones. In recent years, bats have gained significant notoriety for being implicated in numerous emerging infectious disease events, and their importance as reservoir hosts for viruses that can cross species barriers to infect humans and other domestic and wild mammals is increasingly recognized. Historically, there has been a significant body of work on the role of bats as reservoirs of infectious agents, but relatively little available information on members of the suborder Megachiroptera (flying foxes and fruit bats). Although the role of bats in harbouring viruses (alphaviruses, flaviviruses, rhabdoviruses and arenaviruses) is well established, there is increasing global interest in evaluating the broad range of potential infectious agents that bats harbour, with a particular focus on potential emerging pandemic threats. This concern may be somewhat exaggerated, as bats themselves do not represent the real threat to people as regards potential pathogens leading to large-scale zoonotic disease events. However, it is worth investigating the infectious agents like noroviruses harboured by bats, and integrating this information with an understanding of the risk of transmission from bats to people or livestock, which may serve as intermediate hosts and transmission vectors linking bats and people. In this study, Norovirus was detected in two samples (2/20) representing a rate of 10% in bats of the studied area. This finding indicates that bats may be a potential route of transmission of enteric norovirus infection to humans. Viral host switching is probably determined by the chances of interspecies contact, as well as by the concentration and prevalence of virus in the donor species. To judge zoonotic risks associated with bats, when and where these 2 variables would favor transmission must be determined [47-52].
The samples from the location mentioned in our study were selected because they are regularly domiciled with bats and thus provide a certain chance of detection. Attempts to characterize virus dynamics in bat populations have been made earlier by using the examples of lyssaviruses (rabies virus and related species), filoviruses (Ebola and Marburg viruses) and henipaviruses (Hendra and Nipah viruses). However, because these viruses are found rarely, only vague conclusions have so far been made. For instance, increased contact between bats and humans through bat migratory events or fruit harvesting periods have been temporally linked with individual human cases of Ebola and Nipah virus infection. This present study shows the presence of Norovirus in fecal bat from Epinmi, Ondo state. Bats are hosts to a range of zoonotic and potentially zoonotic pathogens. They differ from other disease reservoirs because of their unique and diverse lifestyles, including their ability to fly, often highly gregarious social structures, long life spans, and low fecundity rates. They represent a potential epidemiologic of several diseases that can be fatal to humans, including rabies, Ebola, leptospirosis, histoplasmosis, and pseudotuberculosis [7, 53]. Bats are reservoirs of several pathogens, whose spread may be related to physiological stress associated with habitat loss or alteration [54]. On basis of this, our study shows that the bacterial isolates from bat feacal droppings had multiple antibiotic resistance to antibiotics provided. Levofloxacin and Ampiclox have high susceptibility effect against Gram positive bacteria. Similarly, Ofloxacin and Nalidixic acid having high susceptibility effect against Gram negative bacteria (Table 6a and b). This corroborates with reports of Wibbelt et al. and Calisher, et al. who demonstrated the presence of various pathogenic organisms including bacterial species from bat sources. Human activities that increase exposure to bats will likely increase the opportunity for infections [7, 53]. Like bird droppings, bat guano can contain a potentially infectious fungus Histoplasma capsulatum that causes lung infection known as histoplasmosis [55]. Bats breed in high densities within human populations in some countries, especially in tropics including Nigeria, whereby humans and animals are in close contact. Hence, there is increased risk for cross species transmission. Bat populations appear to be declining noticeably in response to human induced environmental factors like habitat destruction and fragmentation, disturbance to caves, depletion of food resources, overhunting for bush meat and persecution. Other factors that stress the environment such as use of pesticides, infectious disease, and industrial activities including wind energy turbine contributes to this. The role of bat as reservoir of deadly etiologic agent become more pronounced during the COVID-19 pandemic when it was documented as a probable zoonotic source of the disease. Also, bats are known to feed mostly on fruit, therefore this study recommends that fruits bought from markets should be washed thoroughly before consumption. Nevertheless, much caution should be taken on bat hibernating in domestic places as well as the humans and animals contact. The result obtained in this study will help in health management system.
We appreciate the support of Environmental Microbiology and Virology research unit of the Department of Microbiology, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria. This is in terms of samples collection and logistics used for the study.
